

Given refinements being experienced in analytical methods and software tools, the multiparent populations will be the resource of choice to undertake genome wide association studies (GWAS), multiparent MARS, and genomic selection (GS). Furthermore, rapid generation of genome-wide marker data coupled with easy access to precise and accurate phenotypic screens enable large-scale exploitation of LD not only to discover novel QTLs via whole genome association scans but also to practise genomic estimated breeding value (GEBV)-based selection of genotypes. Additionally, the rising molecular breeding approaches like marker assisted recurrent selection (MARS) that are able to harness several QTLs are of particular importance in obtaining a “designed” genotype carrying the most desirable combinations of favourable alleles. Availability of second generation mapping populations encompassing multiple alleles, multiple traits, and extensive recombination events is radically changing the phenomenon of classical QTL mapping. Complementing the technological developments, conceptual shifts have also been witnessed in designing experimental populations. We also show how the power to detect a QTL and the mapping accuracy vary, depending on QTL location.Next generation sequencing platforms and high-throughput genotyping assays have remarkably expedited the pace of development of genomic tools and resources for several crops. We show by simulation that QTL explaining 10 % of the phenotypic variance will be detected in most situations with an average mapping error of about 300 kb, and that if the number of lines were doubled the mapping error would be under 200 kb.
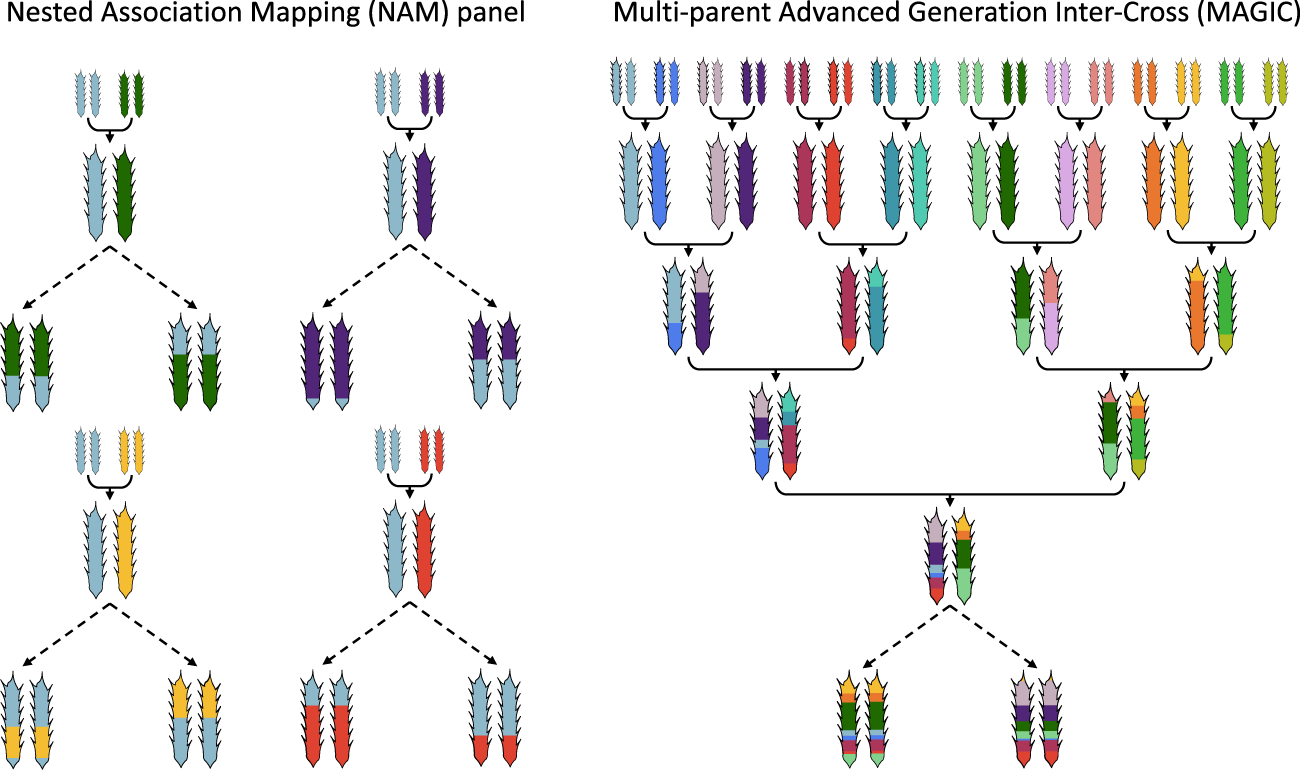
Analytical methods were developed to fine-map quantitative trait loci (QTL) in the MAGIC lines by reconstructing the genome of each line as a mosaic of the founders. These lines and the 19 founders were genotyped with 1,260 single nucleotide polymorphisms and phenotyped for development-related traits. Here, we present the first panel of MAGIC lines developed: a set of 527 recombinant inbred lines (RILs) descended from a heterogeneous stock of 19 intermated accessions of the plant Arabidopsis thaliana. This approach is expected to improve the precision with which QTL can be mapped, improving the outlook for QTL cloning. Here we describe one such alternative, the Multiparent Advanced Generation Inter-Cross (MAGIC). Both of these approaches have some limitations, therefore alternative resources for the genetic dissection of complex traits continue to be sought. Most studies have employed either simple synthetic populations with restricted allelic variation or performed association mapping on a sample of naturally occurring haplotypes. Identifying natural allelic variation that underlies quantitative trait variation remains a fundamental problem in genetics.


 0 kommentar(er)
0 kommentar(er)
